Microbial secondary metabolites have been exploited by mankind for decades, especially as medical drugs. They are complex molecules often produced by biosynthetic genes clusters (BGCs). Despite their enormous societal importance, the ecological functions of microbial secondary metabolites are poorly understood and their roles and impact in natural microbial niches only partially understood. It remains a central question why microorganisms acquire and maintain genes encoding for production of such complex molecules.
Our hypothesis is that microbial secondary metabolites display a much broader spectrum of functionalities than the predominant thinking that these molecules protect the producer from competitors. We believe that secondary metabolites are essential in shaping microbial growth, metabolism and population dynamics at community level and impact ecology and evolution on a grand scale. Furthermore, the production of secondary metabolites itself is shaped by the microbial community and this necessitates the study of natural environments and controlled model systems to elucidate function, diversity, and evolution of these molecules.
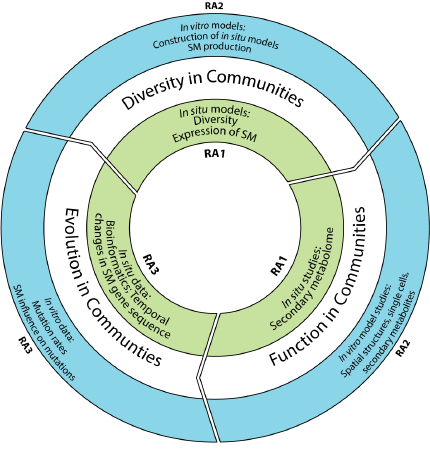
Main Research Areas:
- Natural systems: Microbial diversity and secondary metabolite potential in situ (Diversity in Communities) RA 1.
- Microcosms systems: Influence of secondary metabolites on microbial diversity (Function in Communities) RA 2.
- Diversity and evolution of secondary metabolites (Evolution in Communities) RA 3.